Common Statistics Functions🔗
This guide will walk you through some of the commonly used functions and workflows in Apheris Statistics. It assumes you've already worked through Simulating and Running Statistics Workflows, and are familiar with the basic workflow. If you haven't already, please work through that tutorial before starting this one.
First, import apheris and login to the environment:
import apheris
from aphcli.api import datasets
from apheris_stats.simple_stats.util import SimpleStatsSession, provision
from apheris_preprocessing import FederatedDataFrame
from apheris_stats import simple_stats
apheris.login()
Logging in to your company account...
Apheris:
Authenticating with Apheris Cloud Platform...
Please continue the authorization process in your browser.
Take another look at the datasets, which are available to you:
datasets.list_datasets()
Output:
+-----+-------------------------+--------------+---------------------+
| idx | dataset_id | organization | data custodian |
+-----+-------------------------+--------------+---------------------+
| 0 | whas2_gateway-2_org-2 | Org 2 | Orsino Hoek |
| 1 | whas1_gateway-1_org-1 | Org 1 | Agathe McFarland |
| ... | ... | ... | ... |
+-----+-------------------------+--------------+---------------------+
In this tutorial you will again work on the datasets whas1_gateway-1_org-1 and whas2_gateway-2_org-2.
As a reminder, these are synthetically generated datasets that contain medical data. Both are CSV files and have same structure. One resides on the Gateway of Organization "Org 1" and the other is with "Org 2".
simple_stats_session = provision(
dataset_ids=[
"whas1_gateway-1_org-1", "whas2_gateway-2_org-2"
]
)
simple_stats_session
compute_spec_id: 7b6d7e19-a364-4c2e-85d1-2038d4a00101
Successfully activated ComputeSpec!
SimpleStatsSession(compute_spec_id='7b6d7e19-a364-4c2e-85d1-2038d4a00101')
The provisioning returns a SimpleStatsSession object. The compute_spec_id is an ID that refers to the cluster that has just been activated.
If you wanted to reuse an existing activated Compute Spec with a new Apheris Statistics script, you can instantiate a new session with it:
my_compute_spec_id = simple_stats_session.compute_spec_id
my_second_session = SimpleStatsSession(my_compute_spec_id)
my_second_session
SimpleStatsSession(compute_spec_id='7b6d7e19-a364-4c2e-85d1-2038d4a00101')
Now, create a FederatedDataFrame on the first dataset; whas1_gateway-1_org-1 and another for the second; whas2_gateway-2_org-2:
dataset_ids = ["whas1_gateway-1_org-1", "whas2_gateway-2_org-2"]
fdf_whas1 = FederatedDataFrame(dataset_ids[0])
fdf_whas2 = FederatedDataFrame(dataset_ids[1])
You can investigate the structure of a dataset using the describe function. This will tell you some meta information about the dataset, but without revealing the actual source data.
Run describe on the FederatedDataFrame for the whas1_gateway-1_org-1 dataset to see what columns are available and some basic statistics:
description = simple_stats.describe(fdf_whas1, simple_stats_session)
description[dataset_ids[0]]["results"]
| type | count | count_nan | num_unique_values | percentile_5th | percentile_95th | |
|---|---|---|---|---|---|---|
| afb | float64 | 100 | 0 | 2 | 0 | 1 |
| age | float64 | 100 | 0 | 45 | 48 | 89 |
| av3 | float64 | 100 | 0 | 2 | 0 | 0 |
| bmi | float64 | 100 | 0 | 95 | 18.6 | 35.3 |
| chf | float64 | 100 | 0 | 2 | 0 | 1 |
| cvd | float64 | 100 | 0 | 2 | 0 | 1 |
| diasbp | float64 | 100 | 0 | 54 | 49 | 116.2 |
| gender | float64 | 100 | 0 | 2 | 0 | 1 |
| hr | float64 | 100 | 0 | 60 | 49.8 | 133.1 |
| los | float64 | 100 | 0 | 19 | 2 | 16 |
| miord | float64 | 100 | 0 | 2 | 0 | 1 |
| mitype | float64 | 100 | 0 | 2 | 0 | 1 |
| sho | float64 | 100 | 0 | 2 | 0 | 1 |
| sysbp | float64 | 100 | 0 | 64 | 97 | 200.3 |
| fstat | bool | 100 | 0 | 2 | NaN | NaN |
| lenfol | float64 | 100 | 0 | 91 | 3 | 2175.2 |
Running statistical operations🔗
The first statistics function you'll use here is the count_null function, which counts the number of null values in a column of a dataset. In this example, you will provide one of the FederatedDataFrames from above to run on that dataset:
result = simple_stats.count_null(
fdf_whas1,
column_names=["age"],
aggregation=True,
session=simple_stats_session
)
result
| | |count_null|
|:----|:----|:----|
|total|NA values|0|
|not NA values|100|
This time, do the same, but pass both the FederatedDataFrames from above. This means your operation will run on multiple Gateways at the same time.
You'll see aggregation=True is set; which means that the results from each Gateway will be aggregated prior to returning them to you.
result = simple_stats.count_null(
[fdf_whas1, fdf_whas2],
column_names=["age"],
aggregation=True,
session=simple_stats_session
)
result
| | |count_null|
|:----|:----|:----|
|total|NA values|0|
|not NA values|480|
Now switch aggregation off to see the per dataset results:
result = simple_stats.count_null(
[fdf_whas1, fdf_whas2],
column_names=["age"],
aggregation=False,
session=simple_stats_session
)
result
{'whas1_gateway-1_org-1': {'results': count_null count
total age NA values 0 100
not NA values 100 100},
'whas2_gateway-2_org-2': {'results': count_null count
total age NA values 0 380
not NA values 380 380}}
You can see a full list of statistics functions in the Statistics Reference. You'll see many of the functions are used in a similar way; for example we can use mean_column instead of count_null to calculate the mean age over the data:
result = simple_stats.mean_column(
[fdf_whas1, fdf_whas2],
column_names=["age"],
aggregation=True,
session=simple_stats_session
)
result
69.58333333333334
You can group results by other columns to get more insight into the data. Here we'll use gender, which is categorised numerically as 0 or 1 in this dataset:
result = simple_stats.mean_column(
[fdf_whas1, fdf_whas2],
column_names=["age"],
group_by="gender",
aggregation=True,
session=simple_stats_session
)
result
| |mean_column|
|:----|:----|
|0.0|66.193772|
|1.0|74.712042|
|total|69.583333|
Advanced example: Summary statistics🔗
This more advanced example will demonstrate how to use the pre-processing functionality in the FederatedDataFrame to manipulate the input data in preparation for analysis. You'll then use the tableone function to capture a number of summary statistics about the data.
def prepare_dates(federated_df):
for col in ["sysbp"]:
federated_df.loc[:,col] = federated_df.loc[:,col].to_datetime(unit='M')
for col in ["fstat", "gender", "age"]:
federated_df.loc[:,col] = federated_df.loc[:,col].astype("int")
federated_df["sysbpyr"] = federated_df["sysbp"].dt.year
return federated_df
all_datasets = [fdf_whas1, fdf_whas2]
prepped_datasets = [prepare_dates(d) for d in all_datasets]
numerical_columns = ["hr", "los", "sysbpyr", "fstat", "gender"]
table_one_result = simple_stats.tableone(
session=simple_stats_session,
datasets=prepped_datasets,
numerical_columns=numerical_columns,
numerical_nonnormal_columns=numerical_columns
)
table_one_result
| |col|category|n|mean|std|min|quartile_1|median|quartile_3|max|
|:----|:----|:----|:----|:----|:----|:----|:----|:----|:----|:----|
|0|fstat|total|480|0.422917|0.494022|0.0|0.01|0.01|0.99|1.0|
|1|los|total|480|6.185417|4.767183|0.0|2.82|4.70|7.99|47.0|
|2|sysbpyr|total|480|1981.495833|2.681725|1974.0|1979.92|1980.88|1982.96|1990.0|
|3|hr|total|480|86.660417|23.661134|35.0|68.22|84.83|99.93|186.0|
|4|gender|total|480|0.397917|0.489468|0.0|0.01|0.01|0.99|1.0|
Advanced example: Kaplan-Meier statistics🔗
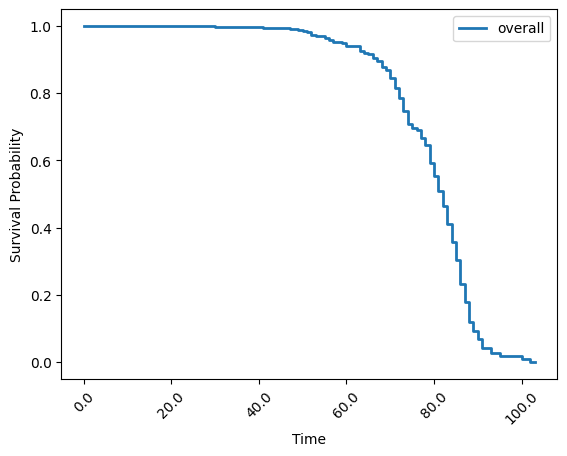
In this final example, you will use Kaplan-Meier to estimate a survival function, measuring the likelihood of the occurrence of a certain event over time.
This example uses a combination of FederatedDataFrame pre-processing and the kaplan_meier function from Apheris Statistics to calculate the likelihood of survival of a patient with respect to some event fstat.
kaplan_meier_result = simple_stats.kaplan_meier(
session=simple_stats_session,
datasets=prepped_datasets,
duration_column_name="age",
event_column_name= "fstat",
stepsize=1,
plot = True
)
The plot that was created by simple_stats.kaplan_meier gives us a first simple visualization. As you have the raw data kaplan_meier_result, you could use your own plotting tools to generate similar figures.
kaplan_meier_result
{'overall': survival_function cumulative_distribution
total (-0.001, 1.0] 1.000000 0.000000
(1.0, 2.0] 1.000000 0.000000
(2.0, 3.0] 1.000000 0.000000
(3.0, 4.0] 1.000000 0.000000
(4.0, 5.0] 1.000000 0.000000
... ... ...
(99.0, 100.0] 0.017072 0.982928
(100.0, 101.0] 0.017072 0.982928
(101.0, 102.0] 0.008536 0.991464
(102.0, 103.0] 0.008536 0.991464
(103.0, 104.0] 0.000000 1.000000
[104 rows x 2 columns]}
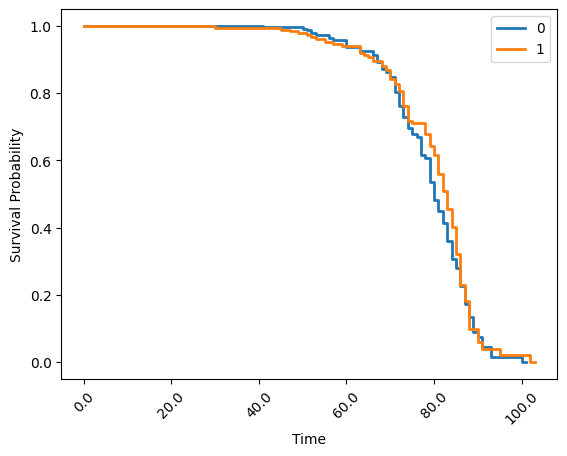
Finally, you can use groupings on kaplan_meier to visualise data by some category. In this case, use the gender column to group the analysis above:
kaplan_meier_result = simple_stats.kaplan_meier(
session=simple_stats_session,
datasets=prepped_datasets,
duration_column_name="age",
event_column_name= "fstat",
group_by="gender",
stepsize=1,
plot = True
)
Close the session🔗
When we are finished, we close the session. This deactivates the deployed cluster, freeing up resources and reducing infrastructure costs.
simple_stats_session.close()
Summary🔗
In this tutorial, you have walked through some of the key statistics functions available to you and see how to use groupings to further explore the data. You have experimented with tableone analysis and visualised survival statistics using Kaplan-Meier.
For more information on the functions you can use with Apheris Statistics, see the Statistics Reference document